1ST WEEK
1. we learnt how to get the protein sequence from different organism. This can be performed from website of MEROPS, search based on domain. Detection of domain is important towards the it’s function.The examples of domain are Clpp and Lon. Once the organism being chosen, the protein sequence will be appeared. From the protein sequence, we can get the peptidase unit and the active site.
2.Then, move to website of National Centre of BioInformatics (NCBI)
NCBI::::> Sequence analysis::::>Protein Blast::::>BlastP
-fill the blank with protein sequence of organism
- click non redundant on database
- then click psi.blast before clicking BLAST
From the blast, we can get the domain, description and identity. Description and identity can be found on the allignment. Identity means that how much the similarity. In short, we do BLAST to search the similarity.
8
2ND WEEK
On 2nd week, Clustalx and Artemis had been used. Clustalx is meant for clustering all those protein sequences while Artemis is a sequence viewer and annotation tool.
1.Clustalx software
a.Wordwrap sequence in notepad
b. Clustalx:::>file:::>load sequence:::>choose sequence from file
complete alignment in order to aligned all the respective sequence.
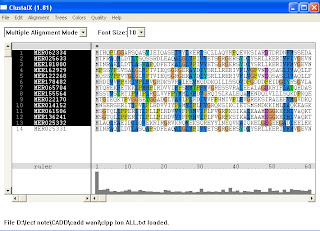
≈ output : dnd (dendogram)…………..need TREEVIEW to view
aln (align all the sequence)………..can b viewed by micr.word also
~ Treeview is used to view the hierarcy of organism
2. Alignment:::>complete alignment:::> align
3. Website MEROPS:>organism:::>eg. Bulkholderia pseudomallei
-choose SJ –S16 (*SJ – clan, S16- family)…for Lon
-copy sequence in notepad
-repeat procedure for SK-S14…….for Clpp
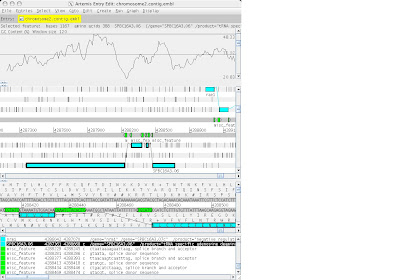
4. Artemis v5:::>file::>open:::>BPs.dbs:::>choose certain sequence from notepad clpplonbp
~ Go to:::>navigator:::>paste that certain sequence in blank at find aa string
~ create:::>mark ORF in range
~ output appeared 3 sequence …..delete sequences without similarity
1 feature left: edit:::>trim selected feature to met
5.To find another sequence which may have similarity within the previous sequence
a) repeat all in step 4
b)CDS:::>edit::>edit selected feature:::>copy name from notepad eg. MER165606
c)paste and replace “none”…apply…ok
6. Website NCBI
a)BLAST:::>protein blast:::>copy paste sequence:::>choose database : pdb
b)Alignment 1st line: 1TYFA (* 1TYFA – ID of pdB):::>Open 1TYF
7. open website Protein Data Bank::::>search IRR9:::>download file:::>save FASTA, save pdb
3RD WEEK
PDB is for homology modeling where the amino acid sequence of a specific protein (target) is known, and the 3-D structures of proteins related to the target (templates) are known.
Eg . Clpp …..1TYF.pdb
Lon…….1RR9.pdb
Open file with RasWin…..~output: 3D structure
3D structure info: eg. Green color: helix, Yellow color: B-shape (straight)
only until here I can recall...plz correct me if there is mistake..





